Today, we will look in more detail into convergence of Markov chains – what does it actually mean and how can we tell, given the transition matrix of a Markov chain on a finite state space, whether it actually converges.
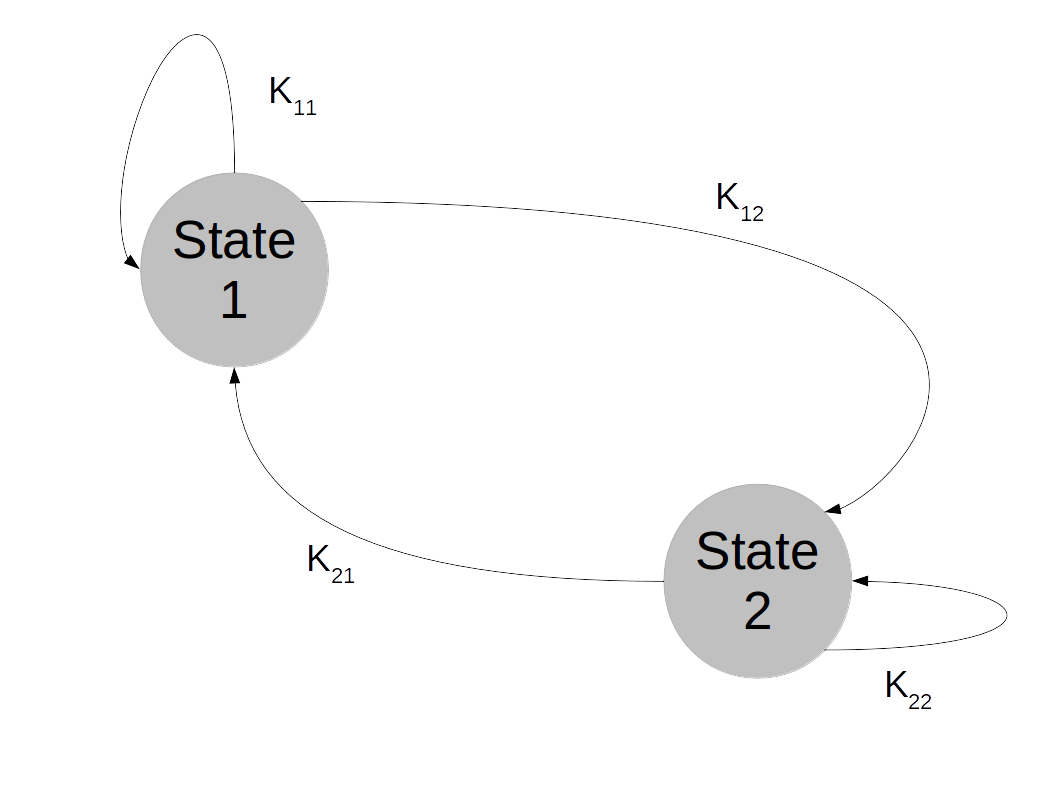
So suppose that we are given a Markov chain on a finite state space, with transition probabilities described by a matrix K as in the previous post on this topic.
We have seen that the distribution of Xn is given by where
is the initial distribution. So if Kn converges, we can expect the distribution of Xn to converge as well. If there is a matrix
such that
then of course
In other words, each row of
will have the property that
. Interpreting K as transition probabilities, this implies that if the distribution of Xn is
, the distribution of Xn+1 will again be
. Any distribution, described by a row vector
with row sum 1, for which this holds is called an invariant distribution and traditionally denoted by
. In other words, invariant distributions correspond to eigenvectors of the transposed matrix KT with eigenvalue 1, and our argument has shown that if
converges to
, then every row of
will be an invariant distribution.
So convergence implies the existence of an invariant distribution. Let us next try to understand whether this invariant distribution is unique. There are obvious examples where this is not the case, the most trivial one being the unit matrix
Obviously and the chain converges, but every vector with row sum one is an invariant distribution. This chain is too rigid, because in whatever state we start, we will stay in this state forever. It turns out that in order to ensure uniquess of an invariant distribution, we need a certain property that makes sure that the states can move around freely which is called irreducibility.
Intuitively, we say that a Markov chain is irreducible if any state can be reached from any other state in finitely many steps. For a finite state space, this is rather easy to formalize. A finite Markov chain is called irreducible if, given two states i and j, we can find some power n such that . In other words, given a row index i and a column index j, we can find a power n such that the element of Kn at (i,j) is not zero. It turns out that if a chain is not irreducible, we can split the state space into smaller areas that the chain – once it has entered one of them – does not leave again and on which it is irreducible. So irreducible Markov chains are the buildings blocks of more general Markov chains, and the study of many properties of Markov chains can be reduced to the irreducible case.
Now let us assume that our chain is in fact irreducible. What else do we have to ask for to make sure that it converges? Again, let us as look at an obvious example.
All even powers of this matrix are the unit matrix and all odd powers are K itself, so the Markov chain described by this transition matrix is clearly irreducible. However, there is still something wrong. If, for instance, we are in state 1 at the time t, then the probability to still be in state 1 at time t+1 is zero. Thus we are actually forced to move to state 2. In a certain sense, there is still an additional constraint on the the behaviour of the chain – the state space can be split in two pieces and we cycle through these pieces with every step. Such a chain is called periodic. If a chain is periodic, it is again obvious that we can think of it as two different chains, one chain given by the random variables X0, X2,… at even times and the other chain given by the random variables X1, X3, … at odd times. Thus again, there is a way to split the chain into parts.
So let us now focus on chains that are irreducible and aperiodic. Can we tell whether the chain converges? The answer is surprisingly simple and one of the most powerful results in the theory or Markov chains: every finite Markov chain which is irreducible and aperiodic converges. Thus once we have verified aperiodicity and irreducibility, we can be sure that the limit exists. We can say even more – we have seen that the rows of the limit are invariant distributions for K. However, one can show that an irreducible Markov chain can only have one invariant distribution. Thus we can conclude that all rows of the matrix
are identical. In fact, they are all equal to the (unique) invariant distribution that again corresponds to an eigenvector of KT with eigenvalue one.
This gives us two different approaches to calculating the invariant distribution and the limit . First, we can form high powers Kn to approximate the limit
. Or, alternatively, we can search for eigenvectors of the transposed matrix KT with eigenvalue 1 using one of the known algorithms to calculate eigenvectors and eigenvalues.
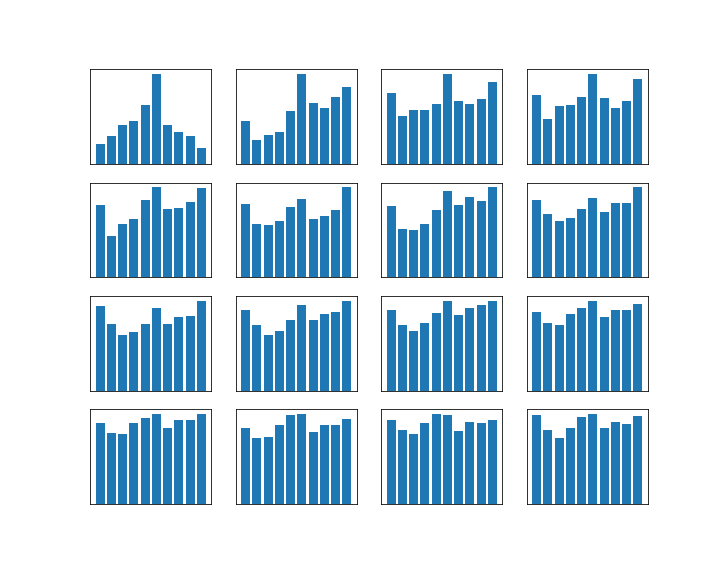
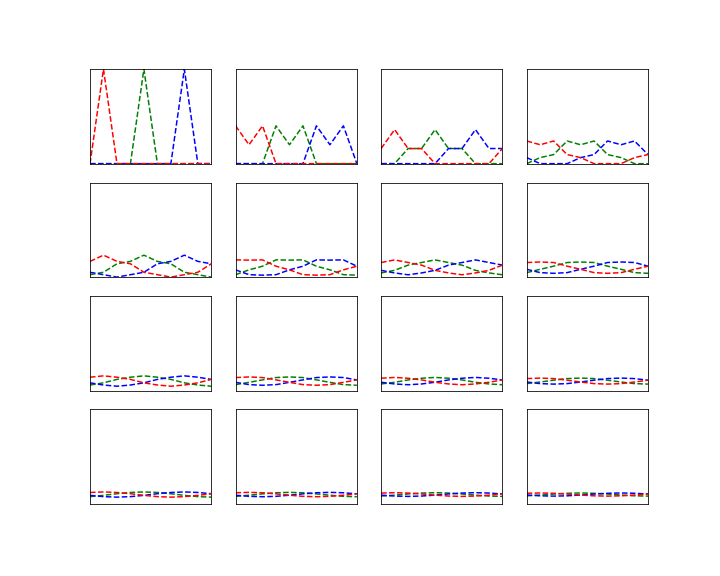
Let us do this for the example of the random walk on a circle that we have studied earlier. For simplicity, we will use N = 4 this time. The transition matrix with p = 0.8 is
We can then easily implement both approaches in Python using the numpy library.
So we see that the approach to take the eigenvalues and eigenvectors gives us – in this case – the exact result (and a direct computation confirms that this is really an eigenvector), whereas the matrix power with n=20 gives a decent approximation. Both results confirm what we have observed visually in the last post – the distribution converges, and it does in fact converge to the uniform distribution. So roughly speaking, after sufficiently many steps, we end up everyhwere in the state space with equal probability.
So far, we have only discussed the case of a finite state space. In the general case, the situation is more complicated. First, we need a better definition of irreducibility, as on a general state space, the probability measure of a single point tends to be zero. It turns out that the solution is to define irreducibility with respect to a measure on the state space, which is called -irreducibility. Once we have that, we find two potential behaviours that do not show up in the finite case.
First, it might happen that even though the chain is irreducible, it does not properly cycle through the state space and revisits every point with finite probability, but – intuitively speaking – heads off to infinity. Such a chain is called transient and does not converge.
Second, even if the chain is not transient, we might be able to find an invariant distribution (a measure in this case), but this measure might not be a probability distribution as it is not finite. Chains with this property are called null recurrent.
And finally, the concept of converge needs to be made more precise, and it turns out to have a subtle dependency on the starting point which can be fixed by assuming so called Harris recurrence. But all this can be done and delivers a very similar result – a convergence theorem for a large class of chains (in this case aperiodic Harris recurrent Markov chains). If you would like to get deeper into this and also see proofs for the claims made in this post and references, I invite you to take a look at my short introduction to Markov chains.
We close this post with one remark which, however, is of crucial importance for applications. Suppose we have a convergent Markov chain Xt, and a function f on the state space. If the Xt were independent and identically distributed, we could approximate the expectation value of f as
where dP is the (common) distribution of the Xt. For Markov chains, however, we know that the Xt are not independent – the whole point of having a Markov chain is that Xt+1 does actually depend on Xt. However, in our example above, we have seen that the dependency gets trivial for large t as in this example, all entries in the transition matrix become equal in the limit.
Of course this is a special case, but it turns out that even in general, the approximation of the expectation value by averages across the sample is still possible. Intuitively, this is not so surprising at all. If the distributions of the $X_n$ converge to some invariant measure
, the
will, for large n, be approximately identically distributed, namely according to
. Moreover, heuristically we have for large n, m and a fixed starting point s:
On the other hand, this product is again approximately the product
Thus, intuitively, Xn and Xm are almost independent if n and m are large, which makes us optimistic that the law of large numbers could actually hold.
In fact, if Xt converges (again I refer to my notes for a more precise definition of convergence in this case), it turns out that the law of large numbers remains valid if we integrate f with respect to the invariant distribution :
This is extremely useful in applications, where often the primary purpose is not so much to obtain a sample, but to approximate otherwise intractable integrals! Once we have found a Markov chain that converges to the invariant distribution , we can calculate integrals over
by running a long Markov chain until converge and then taking the average value of the function that we want to integrate for a large number of subsequent points from this chain. In fact, this is how most applications work – and this is more or less what we also did when we applied Markov chains to the problem of calculating gradients in the PCD algorithm, where this chain is given by the state of the negative particles in subsequent iterations.
So given a distribution , we are now lead to the question how we can possibly find a Markov chain for which this distribution is invariant and which converges. The most general answer to this question is a class of algorithms known as Metropolis-Hastings algorithms which we study in the next post in this series.